A retrospective study among head and neck cancer patients found that a multivariate RNA biomarker test performed equally as standard PD-L1 immunohistochemistry (IHC) testing for predicting responses to anti-PD1-based immune checkpoint inhibitors. In the study, the Predictive Immune Modeling technique from Cofactor Genomics detected an equal number of predicted responders to anti-PD-L1 therapies compared with IHC, or 80 percent. The PIM test resulted in fewer false positives comparatively from 68 percent to 52 percent, respectively. The study was conducted in collaboration with the Washington University School of Medicine in St. Louis and presented at the 2020 Multidisciplinary Head and Neck Cancers Symposium.
The study assessed the predictive response of 107 patients with recurrent or metastatic squamous cell carcinoma of the head and neck previously treated with PD-L1 therapies—either pembrolizumab, nivolumab, or both therapies. Pre-treatment solid tumor tissues were analyzed using PIM compared with an open-label PD-L1 IHC assay.
Head and neck cancer is one of many cancers approved by the FDA for immune checkpoint inhibitors. “The challenge for these immune checkpoint inhibitors is that when they work, they work really well. But they only work, on average, for about 25 percent of the affected population,” says Jarrett Glasscock, CEO of Cofactor Genomics. Predicting who will respond to PD-L1 therapy in advance would spare patients from the harm of pursuing therapies likely to fail.
While the two techniques responded equally in terms of sensitivity, the PIM test reduced the false positive rate by 16 percent. But half of the samples still led to a false positive test.
“Better tools are needed to provide clinicians and patients with a more accurate understanding of tumor response ahead of treatment decisions,” explained Douglas Adkins, M.D., who led the study. “The use of multidimensional biomarkers rather than single analyte biomarkers represents a promising new approach.”
Using an RNA sequencing approach, Cofactor Genomics’ focus is on modeling the RNA or transcriptome of each individual immune cells, including T- and B-cells, macrophages, monocytes, Tregs, and natural killer cells. “These are all immune cells that when in the tumor microenvironment they can be predictive of whether a patient is going to respond to an immunotherapy, or more specifically, an immune checkpoint inhibitor,” says Glasscock.
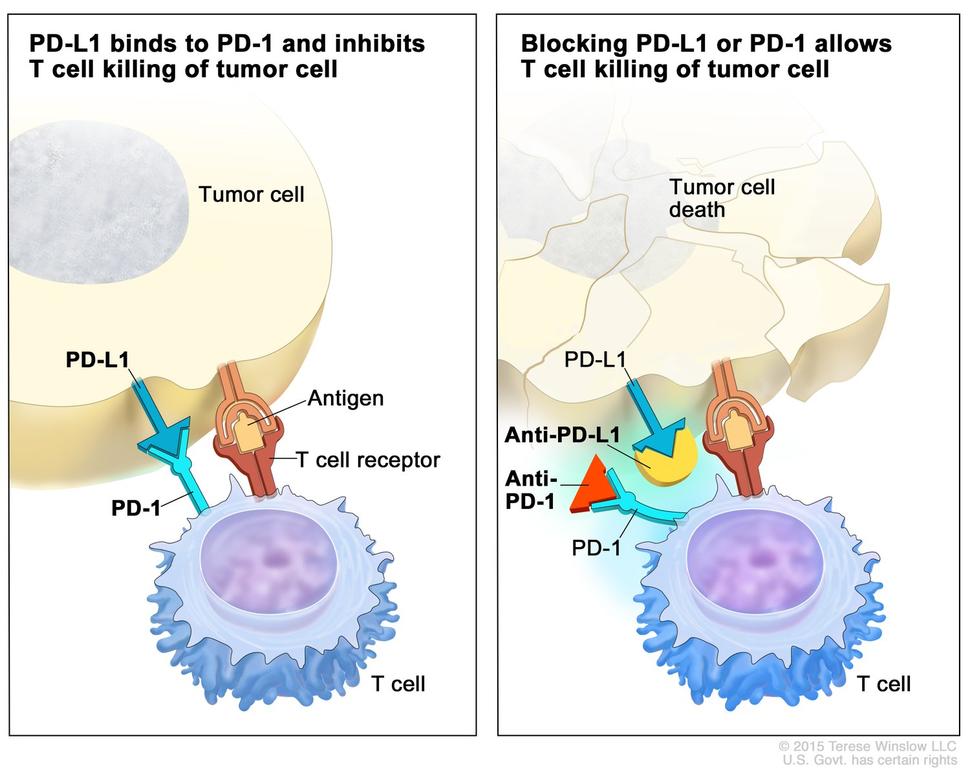
The ImmunoPrism assay is the result of the company’s current state of modeling the RNA of the immune system. “Because of those models we are able to understand the composition of the tumor microenvironment with respect to the immune system,” he says. “The underlying thesis is if you have a more comprehensive biomarker — more than just one analyte like PD-L1 —one hopes to do a better job of predicting who the responders are going to be.”
The company’s method relies on applying machine learning to train results to identify RNA biomarkers in responders and non-responders to immunotherapy.
RNA sequencing results are translated into immune cell percentages that are in the tumor microenvironment.
“The assay includes a combination of immune cell percentages paired with RNA expression of genes — these are typically the immune escape genes that are so important in immune oncology,” says Glasscock. The test also collects information on expression of co-stimulators and co-inhibitors. “It is really a comprehensive view of not only RNA expression but immune cell composition.”
While the false positive rate is still high, specificity is likely to improve over time. “We have taken a very conservative approach in this study, “adds Glasscock. “As the technique becomes more precise, we hope to match on the diagnostic side the excitement we have seen on the treatment side with immunotherapy in the last five years especially.”