Targeted sequencing is an economical way to obtain sequencing reads that are limited only to genomic regions of interest. There are two different approaches used in targeted sequencing—exome capture and amplicon sequencing.1,2 While exome capture has advantages for whole-exome sequencing (30 Mb for the human genome), amplicon libraries offer more specific, cost-effective results in a shorter time and are ideal for use when the targeted regions are smaller than 200 kb.3
In many research and diagnostic scenarios, the sequencing of a limited number of genes is required for a large number of samples. Amplicon libraries are the method of choice in these scenarios.
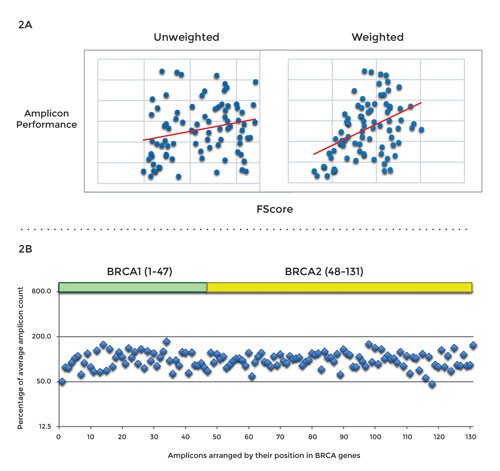
There are two major categories of DNA amplicon libraries—those used for the screening of germline mutations and those used for the screening of somatic mutations. The screening of germline mutations does not require deep sequencing, and high-quality input material typically is available. These panels usually contain a low number of amplicons that, when amplified with high uniformity, allow for a high level of multiplexing and economical sequencing.
Non-uniform coverage in amplicon panels necessitates more reads per panel, which can dramatically increase sequencing costs. Amplicon panels for the screening of somatic mutations require deeper sequencing and often must perform well with partially degraded samples. From an economic standpoint, high uniformity is imperative because the detection of rare mutations (with an abundance below 5%) depends on high coverage.
For the rest of this article, click here.